On Marginal Likelihood and Cross-Validation
Here's a paper someone has pointed me to, along the lines of "everything that works, works because it's Bayesian":
- Edwin Fong, Chris Holmes (2019) On the marginal likelihood and cross-validation
I found this paper to be lacking on the accessibility front, mostly owing to the fact that it is a mixture of two somewhat related but separate things:
- (A) a simple-in-hindsight and cool observation about the relationship between marginal likelihoods and cross validation which I will present in this post, and
- (B) a somewhat tangential sidetrack about a generalized form of Bayesian inference and prequential analysis which I think is mostly there to advertise an otherwise interesting line of research Chris Holmes and colleagues have been working on for some time. The advertising worked for sure, as I found the underlying paper (Bissiri et al, 2016) quite interesting as well. I will leave discussing that to a different post, maybe next week.
The marginal likelihood and cross-validation
To discuss the connection between marginal likelihoods to (Bayesian) cross validation, let's first define what is what.
The marginal likelihood
First of all, we are in the world of exchangeable data, assuming we model a sequence of observations $x_1,\ldots,x_N$ by a probabilistic model which renders them conditionally independent given some global parameter $\theta$. Our model is thus specified by the observation model p(x\vert \theta) and prior $p(\theta)$. The marginal likelhiood is the probability mass this model assigns to a given sequence of observations:
$$
p(x_1,\ldots,x_N) = \int \prod p(x_i \vert \theta) p(\theta) d\theta
$$
Important for the discussion of its connection with cross-validation, the marginal likelhihood, like any multivariate distribution, can be decomposed by the chain rule:
$$
p(x_1,\ldots,x_N) = p(x_1)\prod_{n=1}^{N-1}p(x_{n+1}\vert x_1,\ldots, x_{n})
$$
And, of course, a similar decomposition exists for any arbitrary ordering of the observations $x_n$.
Cross-validation
Another related quantity is a single-fold leave-$P$-out cross-validation. Here, we set the last $P \leq N$ observations aside, fit our model to the first $N-P$ observations, and then we calculate the average predictive log loss on the held-out pounts. This can be written as:
$$
- \sum_{p=1}^{P} \log p(x_{N-p+1}\vert x_1, \ldots, x_{N-P})
$$
Importantly, here, we assume that we perform Bayesian cross-validation of the model. I.e. in this formula, the parameter $\theta$ is integrated out. In fact what we're looking at is:
$$
- \sum_{p=1}^{P} \log \int p(x_{N-P+1}\vert \theta) p(\theta \vert x_1, \ldots, x_{N-P}) d\theta
$$
Now of course, we could leave any other subset of size $P$ of the observations out. If we repeat this process $K$ times with a uniform random subset of datapoints left out each time, and average the results over the $K$ trials, we have $K$-fold leave-$P$-out cross validation. If $K$ is large enough, we might be trying all possible subsets of $P$ with the same probability. I will cheesily call this $\infty$-fold cross-validation. Mathematically, $\infty$-fold leave-$P$-out Bayesian cross-validation is the following quantity:
$$
- \frac{1}{N \choose P} \sum_{\substack{S⊂\{1\ldots N\}\\|S|=P}} \sum_{i \in S} \log p(x_i\vert x_j : j \notin S),
$$
which is Eqn (10) in the paper with slightly different notation.
The connection
The connection I think is best illustrated in the following way. Let's consider three observations, and all the possible ways we can permute them. There are $3(3+1)/2 = 6$ different permutations. For each of these permutations we can decompose the marginal likelihood as a product of conditionals, or equivalently we can write the log marginal likelihood as a sum of logs of the same conditionals. Let's arrange these log conditionals into a table as follows:
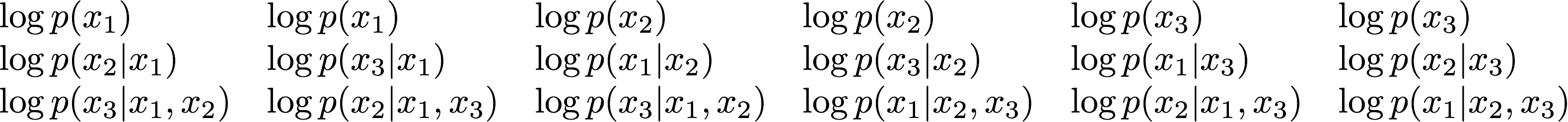
Each column corresponds to a different ordering of variables, and summing up the terms in each column gives the log marginal likelihood. So, the sum of all the terms in this matrix gives the marginal likelihood times 6 (as there are 6 columns). In general it gives $N(N+1)/2$ times the marginal likelihood for $N$ observations. Now look at the sums of the terms in each row. The first row is full of terms you'd see in leave-$3$-out cross validation (which doesn't make too much sense with $3$ observations). In the second row, you see terms for leave-2-out CV. Third row corresponds to leave-1-out CV. So, if you do some careful combinatorics (homework) and count how many duplicate terms you'll find in each row, one can conclude that the sum of leave-K-out $\infty$-fold Bayesian cross-validation errors for all values of $K$ gives you the log marginal likelihood times a constant. Which is the main point of the paper.
This observation gives a really good motivation for using the marginal likelihood, and also gives a new perspective on how it works. For $N$ datapoints, there are 2^N-1 different ways of selecting a non-empty test set and corresponding training set. Calculating the marginal likelihood amounts to evaluating the average predictive score on all of these exponentially many 'folds'.
Hooray?
Before we jump to the conclusion that cross-validation, too, works only because it is essentially an approximation to Bayesian model selection, we must remind ourselves that this connection only holds for Bayesian cross-validation. What this means is that in each fold of cross-validation, we integrate $\theta$ in a Bayesian fashion.
In practice, when cross-validating neural networks, we usually optimize over the parameters rather than integrate in a Bayesian way. Or, at best, we use a variational approximation to the posterior and integrate over that approximately. As the relationship only holds in theory, when exact parameter marginalization is performed, it remains to be seen how useful and robust this connection will prove in potential applications.
